How to find what you need¶
Both pipelines are large, complex networks of schemas, tables, and columns. Here are some general suggestions to help you find what you need. There are further suggestions, specific to each pipeline, in the relevant sections.
Finding a table: the Diagram¶
Datajoint includes a Diagram feature, to display the pipeline as a connected network. This can operate either on a schema object, or a table, or some combination of the two. You may also see the keyword “ERD” used in places - this is a more mathematically correct description (“entity relationship diagram”), but it means the same thing, and executes exactly the same Python code to give you a graphical output.
Electrophysiology: connect and view the ephys diagram
import datajoint as dj
ephys = dj.create_virtual_module("ephys", "group_shared_ephys")
dj.Diagram(ephys)
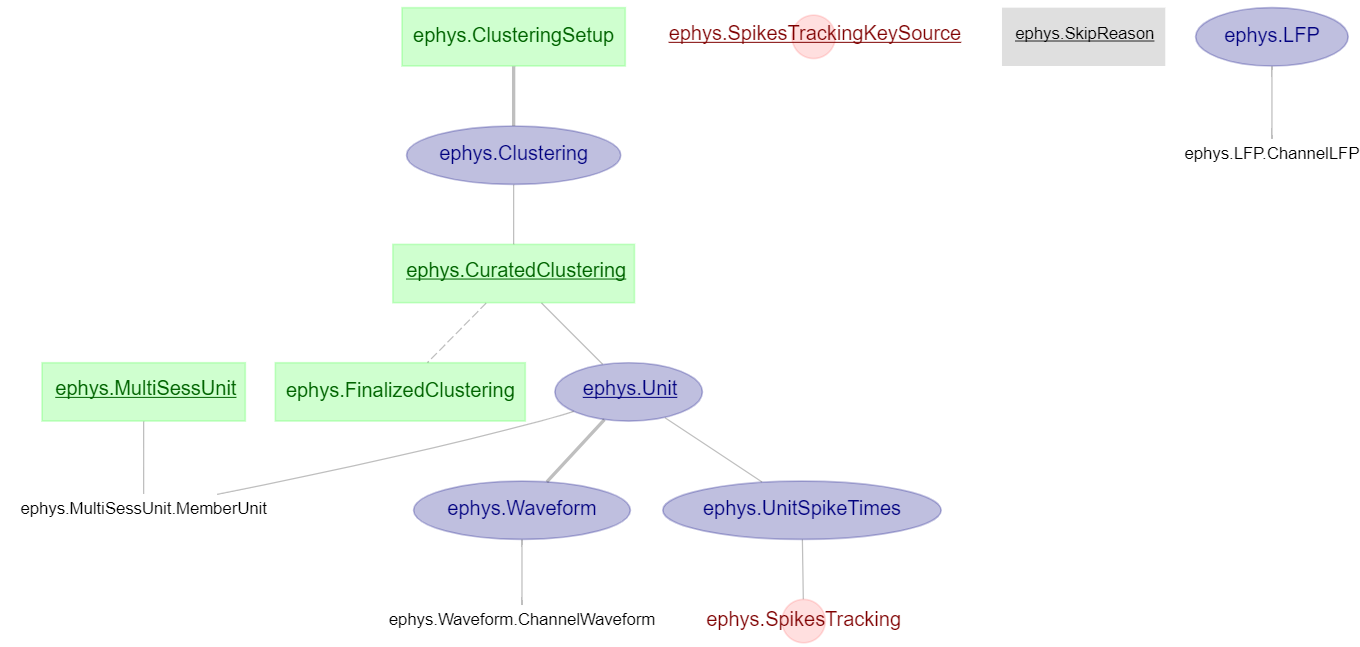
Image showing the entire ephys diagram (note: the ephys pipeline also contains other schemas, such as one for analysis and one for behaviour).
Imaging: connect and view the entire imaging pipeline diagram
import datajoint as dj
imaging = dj.create_virtual_module('imaging', 'group_shared_imaging')
imaging.schema.spawn_missing_classes()
dj.ERD(imaging)
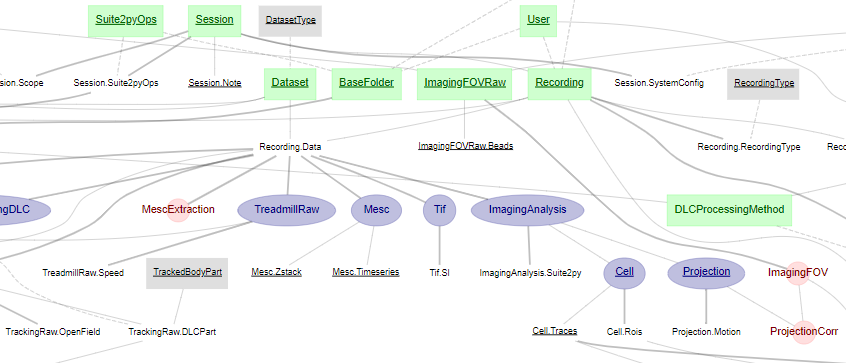
Image showing part of the imaging schema.
What do the colours mean?¶
Examples are from the imaging schema
Grey (lookup) or Green (manual): data gathered from outside the pipeline
Small tables containing general facts, settings or other data entered by hand or with external helper scripts.
E.g.
RecordingTypecontains a list of the types of recordings, e.g. OpenField, Wheel, etc.E.g.
BaseFoldercontains the information provided by the user when ingesting data, such as via the Web GUIPurple (imported): data generated within the pipeline based on data from outside
The content is computed automatically inside the pipeline but access to data outside the pipeline is required to do so.
E.g.
TrackingRawgathers info on time stamps and sampling rate but requires access to the ingested data files to retrieve it.Red (computed): data generated entirely within the pipeline
Data computed automatically entirely inside the pipeline.
E.g.
Suite2pycontains information about Suite2P python jobs that the pipeline has executed.No colour/white (part table): a ‘sub-table’ to the preceding ‘master/parent’ table
Some tables would get very big if all the data computed was put into a single table. Instead, the data is split up/organised into ‘sub-tables’ that are intrinsically linked to their ‘master table’.
E.g.
Cellgathers data from Suite2p, which is very rich. Two part tables are therefore used instead of one, categorising the data intoCell.Traces(dF/F trace etc.) andCell.Rois(number of pixels, ellipticity etc.). Their master tableCellonly contains acell_idfor referencing.E.g.
TuningMapcontains data equivalent to ratemaps in the ephys pipeline, such as the number of fields in an open field recording. But each field within a tuningmap also has a number of values associated with it, such as peak rate. These are saved inTuningMap.Fields. I.e. unlike the example above, these tables do not have the same size: for each tuningmap (one row inTuningMap) there are multiple fields (saved as multiple rows inTuningMap.Fields).
For more info on these table types, check out the Datajoint website.
Viewing smaller sections of the Diagram¶
The Diagram can help you find your way to the right table, although in some cases, the resulting output can be so overwhelmingly large, that it’s just not useful.
You can also search a specific area of the pipeline. If you know about a specific table, and want to know about its “neighbours”, you can use the diagram a bit like an equation, requesting a certain number of neighbours above or below the table. For example:
Ephys
dj.Diagram(ephys.Spikes) +2
Imaging: visualise the table structure two levels on from the Cell table
dj.ERD(Cell)+2
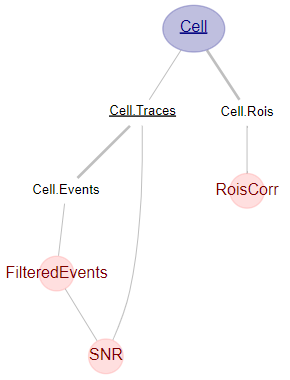
The Cell table has two part tables, Cell.Rois and Cell.Traces. The Cell.Traces table is used by Cell.Events, in which spikes inferred from the traces are stored. These are then filtered in FilteredEvents.
You can also add diagrams together - or subtract - to get a more complex overview that keeps or avoids certain sections of the network
Ephys: view only specified parts of the diagram
analysis = dj.create_virtual_module("analysis", "group_shared_analysis")
dj.Diagram(ephys) + dj.Diagram(analysis) - dj.Diagram(analysis.ShuffledScores)
Imaging: view only specified parts of the diagram
dj.ERD(Recording)+dj.ERD(Cell)+dj.ERD(ImagingAnalysis)
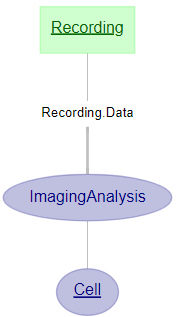
Finding the data you need: columns¶
Having found a relevant table, you need to know if it contains the information you need and if so, how to retrieve it. I.e. you need to know which column name to search for. Here, there are numerous options (Option 1 and Option 3 are perhaps the most useful ones).
Option 1¶
One option is simply to print out a representation of the table itself. This prints out a nicely formatted dataframe, showing all of the columns in that table and their names:
Ephys
ephys.CuratedClustering()
Imaging: check what is in the Cell table
Cell()
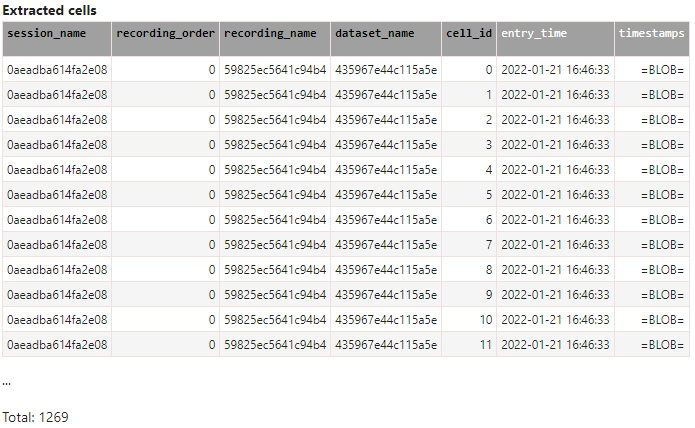
The Cell table contains five primary keys (black font) and one non-primary key (white font) for each and every cell within the database (at the time of writing: >1200 cells).
Option 2¶
Alternatively, you can use one of the helper methods provided by Datajoint. This prints out the text definition that was used to create the table, including any comments that were associated with those columns. However, it can hide some of the column names, as they are implicit in the description, as references to other tables:
Ephys
ephys.CuratedClustering.describe()
Imaging: view a (limited) description of the Cell table
Cell.describe()

The --- separates between primary (above) and non-primary (below) attributes, and can typically be thought of as:
the “things” needed to perform the computation
the results of the computation
Option 3¶
Another alternative, that includes every column name and comment is the heading:
Ephys
ephys.CuratedClustering.heading
Imaging: view every column name of the Cell table
Cell.heading
 Unlike
Unlike .describe(), .heading lists every single column name for the table in question.
Searching for a specific column name¶
Sometimes, you need to go the other way around - you know (or suspect) a column name, but don’t know what table it is in. This is… a bit harder to do, and Datajoint doesn’t yet provide very good tools for it, although development work is on-going
The below function provides a fragile workaround to the problem: if you know the precise name of the column you want, then it will help. It implements exact, case-sensitive matching, so it will not help with more general queries where you do not know the exact name of the column
def search(col_name, *args):
"""Search for the table in which a specific secondary attribute column appears
If the column name features in multiple tables, then all tables (in the provided
schemas) in which that column appears will be listed
Parameters
----------
col_name : str
Precise name of a column that is not a primary key attribute
*args:
One or more schema objects
Returns
-------
list
The table(s) in which that column occurs
Examples
--------
search("grid_score", ephys, analysis, acquisition)
>>> [ephys.analysis.GridScore, ]
search("electrode_config_id", acquistion, ephys)
>>> [ephys.acquisition.Recording, ephys.ephys.FinalizedClustering]
search("fcorr",Cell)
>>> [datajoint.user_tables.Traces]
"""
tables = []
for schema in args:
for key, value in schema.__dict__.items():
if hasattr(value, "describe"):
for attr in value.heading.secondary_attributes:
if col_name.lower() == attr:
tables.append(value)
if len(tables) > 0:
return tables
else:
raise ValueError(f"Column name {col_name} does not match precisely in any table")