Ephys: Fetching data¶
This guide assumes that you understand the basics of fetching data with Datajoint (covered extensively at the Datajoint documentation). It assumes you have created database interface objects as discussed in Accessing the schemas via Python.
Accessing the schemas via Python¶
The Ephys pipeline is split into multiple separate schemas by theme. Each schema requires its own module, either imported from raw Python code or generated from virtual modules. The following code block will generate all the necessary virtual modules
1 import datajoint as dj
2 animal = dj.create_virtual_module('animal', 'prod_mlims_data')
3 reference = dj.create_virtual_module('reference', 'group_shared_reference')
4 acquisition = dj.create_virtual_module('acquisition', 'group_shared_acquisition')
5 tracking = dj.create_virtual_module('tracking', 'group_shared_tracking')
6 behavior = dj.create_virtual_module('behavior', 'group_shared_behavior')
7 ephys = dj.create_virtual_module('ephys', 'group_shared_ephys')
8 analysis = dj.create_virtual_module('analysis', 'group_shared_analysis')
9 analysis_param = dj.create_virtual_module('analysis_param', 'group_shared_analysis_param')
Individual tables can then be accessed as elements of the above modules.
fetch vs fetch1¶
These two functions can catch the unwary out.
.fetch1()requires exactly one responsive row to your query, and will throw an error if there are either 0, or more than 1, responsive rows..fetch()works with any number of responsive rows: 0, 1, or many. It will always return a list of results, even if there is exactly one row to be fetched, and you will have to index the list to get any single result.
Controlling fetching¶
When fetching data, you often want the fetch structured in some particular way. You can do this structuring yourself, in Python or Matlab on your local computer, but several common forms can be specified for the database server to do for you:
fetch ordering: If you don’t specify any particular ordering, then you should expect the results to be returned in a different order every time you fetch them. This can be fixed by
.fetch(... order_by=<column_name> <order>"):ephys.Unit.fetch(order_by="curation_timestamps ASC"). This will fetch all recorded Units, with the members of the oldest curation first (“ascending”). “DESC” can be used for descending order insteadlimit: sometimes, you may have a query with many results, but you don’t need all of them. For very large queries, requesting only as many as you need can save some time by needing to transfer less data to your computer:
ephys.Unit.fetch(limit=10). Only applies to.fetch(), not.fetch1(). Even iflimit=1, still returns a listas_dict: Forces the data to be returned as a list of dictionaries, instead of a sturctured array
ephys.Unit.fetch(as_dict=True). Only applies to.fetch(), not.fetch1()
Core components¶
The pipeline contains a relatively minimal selection of critical tables that all researchers will care about. In addition there are many peripheral tables that handle administrative functions that few or no researchers need to worry about.
The following screenshot includes the major tables in the pre-analysis pipeline. If you are writing your own from-scratch analyses, then it is quite probable that they will diverge from the main pipeline somewhere within this group.
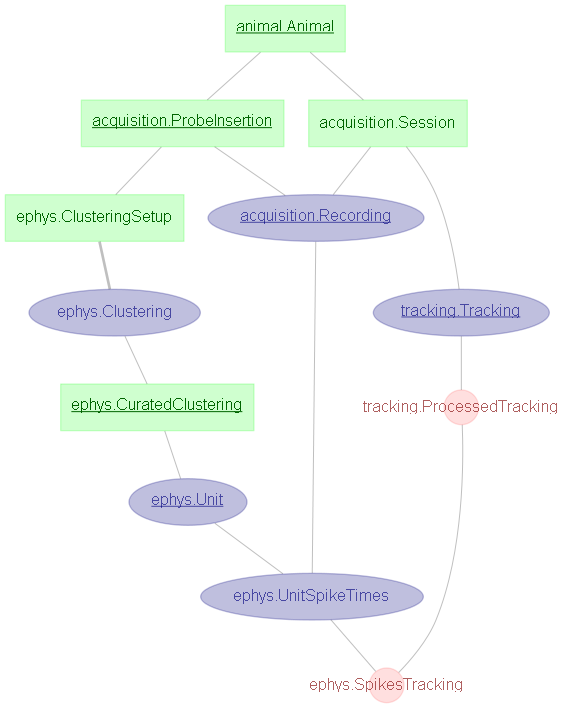
animal¶
The animal schema contains information about the subjects themselves, and is automatically updated from the mLims colony management system. The primary table is animal.Animal, whichcontains the lookup for animal names (e.g. "animal_name = 102").
acquisition¶
The acquisition schema contains information about data acquisition: recording sessions that took place, who was involved, and where the data is stored.
The primary tables are:
acquisition.ProbeInsertion- Contains details on probe implants.acquisition.Sessionand its part tables (.Experimenterfor the researchers involved and.Directoriesfor the data location(s)) - contains details on specific recording sessions that took placeacquisition.Recording- tracks individual recording files (of which there may be several per session)
tracking¶
The tracking schema contains details about tracking the subject’s motion.
ephys¶
The ephys schema contains details about the electrophysiolgical data - Clustering, Units, and matched spike times / tracking
ephys.CuratedClustering- contains details about any user-curated spike-clustering outcomes.ephys.Unit- contains records of each unit in a Clusteringephys.SpikeTimes- contains the time-series of spiketimes per unitephys.SpikesTracking- contains the matched subject motion to spike times.
Restricting Curation by Session¶
From the above image, you can see that Curation is not directly dependent on Session, and so you cannot directly restrict a Curation by a Session. This is because Curations might span across several sessions, and so there is not a one-to-one matching.
1# This will not achieve anything
2# Pick a random session key
3session_key = acquisition.Session.fetch("KEY", limit=1)[0]
4ephys.CuratedClustering & session_key
Instead, you need to use another table to join them up: acquisition.ClusterSessionGroup.GroupMember
1# Instead, you need to include the ClusterSessionGroup table
2# Again, pick a random session key
3session_key = acquisition.Session.fetch("KEY", limit=1)[0]
4ephys.CuratedClustering * acquisition.ClusterSessionGroup.GroupMember & session_key
Tasks and downstream analysis¶
The pipeline has severtal approaches to how a Session is divided. A Session corresponds to a “recording group” - e.g. a researcher goes into his or her lab in the morning, runs a bunch of experiments, and comes out in the afternoon.
Recordings are, roughly, raw data files. E.g. if for some reason a recording starts/stops, then it might be split across several physical files, that all correspond to the same session. Recordings are not a particularly important distinction within the pipeline.
Tasks are the major distinction. They are used to denote separate experimental stages within a session. All downstream analyses are automatically segmented by Task. Tasks are denoted by their start/stop times within a Session, and are completely independent of Recordings.
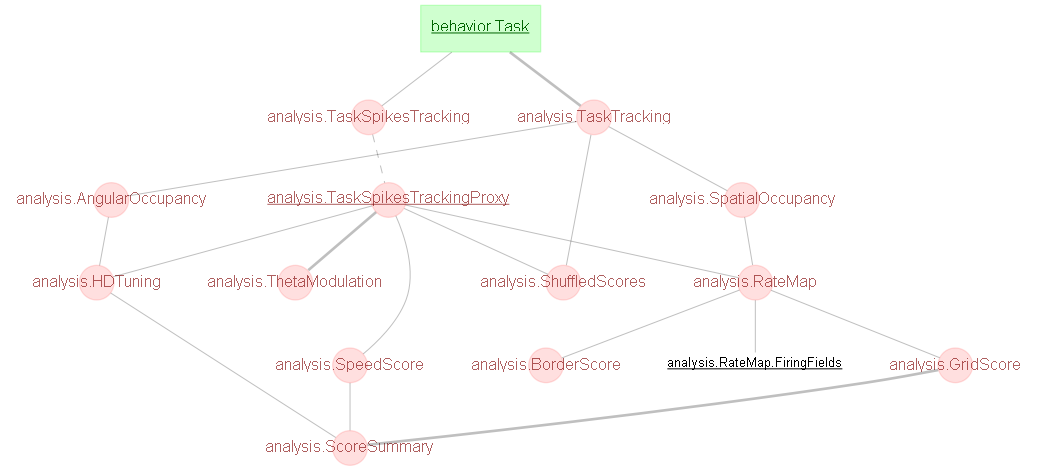
behavior¶
Tasks are handled within the behavior schema.
behavior.Task- contains details of specific Tasks within a single Sessionbehavior.TaskEvent- contains details of other inputs into the acquisition system, e.g. a digital input stream. A single TaskEvent might be, e.g., a list of all photostimulation on/off times.
analysis¶
The two most critical tables are:
analysis.TaskSpikesTracking- stores the contents ofephys.SpikesTracking, but separated by Taskanalysis.TaskTracking- stores the contents oftracking.ProcessedTracking, but separated by Task
All analysis, either included in the pipeline for screening purposes, or your own more advanced analyses, are likely to branch off at, or shortly after, these two tables. Further screening analysis tables are based on the analysis code in both opexebo (docs) and BNT.
In addition, the following tables are provided:
analysis.SpatialOccupancy: the time-map of where the animal spent its time during the sessionanalysis.RateMapandanalysis.RateMap.FiringField: the map of the firing frequency as a function of position, and details on each individual firing field.
TODO
Restricting Analysis by pre-Analysis¶
For technical reasons, tables after analysis.TaskSpikesTracking cannot be directly joined to a set of tables before. In order to replace that link, an administrative table called analysis.TaskSpikesTrackingProxy exists. You should never need to fetch anything from that table, just join with it as part of a query. For example, suppose you wish to find the CuratedClustering associated with a specific Ratemap
1# Pick a random ratemap key
2my_ratemap_key = = analysis.RateMap.fetch("KEY", limit=1)[0]
3ephys.CuratedClustering * analysis.TaskSpikesTrackingProxy & my_ratemap_key
4# should have a single result
If you exclude that * analysis.TaskSpikesTrackingProxy term, then you will get back every CuratedClustering associated with that subject (and that probe if you’re working with multi-probe recordings).
Restricting Analysis by analysis parameter sets¶
Analyses will be run multiple times for as many analysis parameter sets as apply to your username and task types, and you will typically only want to deal with a single one at once. These can either be filtered off on an as-needed basis, but the following can fix things more proactively:
1# Pick a random curation - should hav _many_ TaskSpikesTracking outcomes
2my_curation_key = ephys.CuratedClustering.fetch("KEY", limit=1)[0]
3
4paramsets = (analysis_param.CellAnalysisMethod.CellSelectionParams
5 * analysis_param.CellAnalysisMethod.FieldDetectParams
6 * analysis_param.CellAnalysisMethod.OccupancyParams
7 * analysis_param.CellAnalysisMethod.SmoothingParams
8 * analysis_param.CellAnalysisMethod.ScoreParams
9 * analysis_param.CellAnalysisMethod.ShuffleParams)
10
11# Only evaluate the `default` analysis method
12my_paramset = paramsets & 'cell_analysis_method = "default"'
13analysis.TaskSpikesTracking & my_curation_key & my_paramset